Bioinformatics, biodiversity informatics and data science
People involved
Key research areas
Genomics & Bioinformatics, Biodiversity information systems, Literature mining and data integration to elucidate ecosystem functioning, eDNA Metabarcoding, Omics and Systems Biology bioinformatics pipeline development, containerization and deployment, Data curation and mobilization, Data archaeology and rescue.
 Data is the hard currency by which biodiversity information is exchanged between humans and machines. Biodiversity data are very diverse because they are the product of many disciplines in Biology. Therefore, the communication of biodiversity data in a manner that allows interoperability and data integration is vital and requires major efforts in the adoption of the use of standards and FAIR data principles (Findable, Accessible, Interoperable, Reusable). Our approach involves all levels of biodiversity data science, from data acquisition and curation to the development of interfaces and analytical tools and realisation of FAIR data principles, in an effort to address “Big Data” reality and major scientific and societal challenges.
Data is the hard currency by which biodiversity information is exchanged between humans and machines. Biodiversity data are very diverse because they are the product of many disciplines in Biology. Therefore, the communication of biodiversity data in a manner that allows interoperability and data integration is vital and requires major efforts in the adoption of the use of standards and FAIR data principles (Findable, Accessible, Interoperable, Reusable). Our approach involves all levels of biodiversity data science, from data acquisition and curation to the development of interfaces and analytical tools and realisation of FAIR data principles, in an effort to address “Big Data” reality and major scientific and societal challenges.
Equally, the integration of web services (software available as an online service) requires both their proper annotation and subsequently their composability. Only through these processes the development of proper Virtual Research Environments (VREs) can be achieved, which in turn, makes possible the multi-disciplinary research and production of synthetic knowledge, one of the most urgent scientific and societal needs in current biology.
Systems biology approaches, applied at several levels of biological organization (from molecular to ecosystemic) provide both a theoretical and a technical background for such an integrative analysis to flourish. Besides the aforementioned, literature and web data mining are employed to this end as well as NGS bioinformatics pipeline development, containerization and deployment. eDNA Metabarcoding is a flagship use case.

Genomics & Bioinformatics: the genomic era has progressed heavily based on the rapid development of bioinformatics, the science of developing new ways and algorithms to extract biological knowledge from the tons of biological data produced at a smaller or larger scale worldwide. In IMBBC, we focus on building pipelines that effectively interrogate the genomic information produced in-house or deposited in public databases to gain knowledge on the evolutionary processes taking place across populations, species and communities. We study genomes to understand how they reflect the phenotypic differences among groups, how selection shapes them, how they respond to artificial selection in the case of aquacultured species and how various anthropogenic pressures affect them (e.g. invasions, overexploitation, climate change, sea pollution with organic waste). Our pipelines follow the FAIR principles, are available to the public (e.g., https://genomenerds.her.hcmr.gr/code/) and have been recently been containerized to allow full reproducibility and scalability (e.g. https://github.com/nellieangelova/De-Novo_Transcriptome_Assembly_Pipelines). We have prioritized the development and containerization of pipelines to build whole-genome references of focal species from the sequencing throughput (Illumina & Oxford Nanopore) to the final chromosome-level assembly for a wide range of species (e.g. diatoms, fishes, seagrasses, fungi).
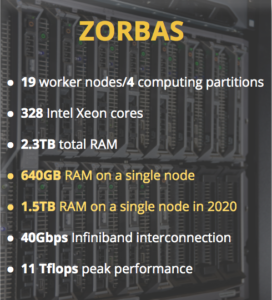
Biodiversity information systems: development, maintenance and continuous support of biodiversity information systems (e.g. national taxonomic backbones, regional and thematic biodiversity databases), platforms, interfaces and data aggregators. The Research Infrastructure LifeWatchGreece includes an ecosystem of e-Services and virtual Labs (vLabs) that aim to support the needs of the scientific community in mobilisation, analysis and sharing of biodiversity datasets, not only for the scientific community, but also for the broader domain of biodiversity management and policy. Ecological analysis and biodiversity indices calculation supported by the infrastructure exceed the capacity of an everyday-use researchers’ laptop or desktop. Analysing up-to global-scale biodiversity datasets is made possible via seamless connection to the underlying High Performance Computing platform
Literature mining and data integration to elucidate ecosystem functioning: ongoing projects gather and mine public knowledge to open new doors in molecular ecology, microbiology and biodiversity and ecosystem research. Large - scale text-mining processes the global scientific literature to recognise organism, environment-type and biological process mentions and extract the statistically significant pairwise associations among them. Linking, for example, microbial taxa with specific biogeochemical cycle reactions reveals processes occurring in a particular environment (such as the bathypelagic water column or the seabed sediment). Metagenetics and metagenomic record metadata are retrieved from global resources and the same type of who-where-what associations are also extracted. Network analysis and visualisation modules bring them who-where-what together to elucidate ecosystem functioning.

EXTRACT highlighting of terms. To quickly identify metadata-relevant pieces of text in a larger document, users may perform a full page tagging. The highlighted entities can indicate candidate segments for subsequent inspection and term extraction. As shown, identified organisms mentions are highlighted in yellow and environment descriptors in orange.
Source: Pafilis E, Buttigieg PL, Ferrell B, Pereira E, Schnetzer J, Arvanitidis C, Jensen LJ. EXTRACT: interactive extraction of environment metadata and term suggestion for metagenomic sample annotation. Database (Oxford). 2016 Feb 20;2016:baw005. (Creative Commons)
The example shows an excerpt of Hurwitz B.L., Sullivan M.B. (2013) The Pacific Ocean virome (POV): a marine viral metagenomic dataset and associated protein clusters for quantitative viral ecology. PLoS One, 8, e57355.
eDNA Metabarcoding, Omics and Systems Biology bioinformatics pipeline development, containerization and deployment: NGS and omics technologies have launched a new era in bio- and eco-assessment. Therefore, a great need for the analysis of their returned data has arisen. The large number of different steps required, the high computational demands and the need to balance between generality and dataset-specific tuning, make this task quite an intriguing one. To address this challenge, we have developed complex bioinformatics pipelines that allow for both robustness, HPC exploitation and user-friendliness. To this end, state-of-the-art technologies are used to develop, implement and share such pipelines. PEMA, a Pipeline for Environmental DNA Metabarcoding Analysis, combines workflow management systems with container-based technologies to illustrate an example of such a pipeline.
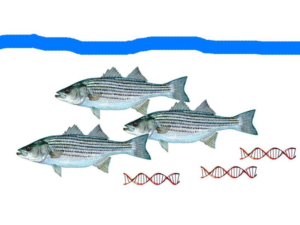 Fish leave bits of DNA behind that researchers can collect. (Mark Stoeckle/Diane Rome Peebles images, CC BY-ND)
Fish leave bits of DNA behind that researchers can collect. (Mark Stoeckle/Diane Rome Peebles images, CC BY-ND)

(source: https://github.com/hariszaf/pema, Haris Zafeiropoulos, Ha Quoc Viet, Katerina Vasileiadou, Antonis Potirakis, Christos Arvanitidis, Pantelis Topalis, Christina Pavloudi, Evangelos Pafilis, PEMA: a flexible Pipeline for Environmental DNA Metabarcoding Analysis of the 16S/18S ribosomal RNA, ITS, and COI marker genes, GigaScience, Volume 9, Issue 3, March 2020, giaa022)
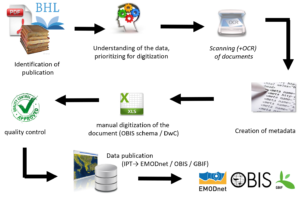
Data curation and mobilization: curation and mobilization of open-access and FAIR datasets concerning aquatic biodiversity and natural history collections are of paramount importance for the establishment of baselines for current and future studies, especially when dealing with biogeographic heterogeneity and global change.
Data archaeology and rescue: rescuing and mobilizing historical marine data from legacy literature and logbooks from historical oceanographic expeditions provide the historical context for present observations, thereby facilitating the process of setting reference conditions for monitoring and management. Linked to this, historical data can be used for reconstructing and modelling past conditions, predicting future trends and shifts in species distribution range, regional species extinctions, biological invasions, and consequences of human activities for the environment and biodiversity. Permanent loss of data has fatal consequences, simply because it is impossible to retrieve those data which were collected from a certain area over a certain period. Consequently, loss of data equals the loss of unique resources and ultimately the loss of our natural wealth.
Related Content
|
Recent Posts
- Seminar: Introduction to hpc and DNA metabarcoding analysis, 7-10 July 2020
- RDA Domain Researchers and Data Experts granted to join the 14th RDA Plenary in Helsinki
- Applying FAIR Principles To Historical Data, Research Data Alliance Blog
- Webinar: Towards ENVRI Community International Winter School DATA FAIRness, July 13 – July 14 2020
- LifeWatch ERIC 2019 Activities Report. The 36-page document is a record of the LifeWatch ERIC’s operations and accomplishments, clarifies what we do and why, and lends greater visibility to our position in the European and global landscape of Research Infrastructures
- EMODnet Biology workshop on data formatting, QC & publishing
-
- The Flanders Marine Institute (VLIZ), the Hellenic Centre for Marine Research (HCMR) and the Istituto Nazionale di Oceanografia e di Geofisica Sperimentale (OGS) have organised a hands-on workshop on data formatting, quality control (QC) & publishing as part of the EMODnet Biology project (phase 3), with the intention to provide training to current and future data providers on how to format and submit their data to the project. The trainers at the workshop were from VLIZ (EurOBIS-EMODnet Bio-LifeWatch Belgium), HCMR (EMODnet Bio-LifeWatch-Greece) & OGS (EMODnet Bio).
































